JGI Plant Gene Atlas
Gene expression across diverse plant species
The JGI Plant Gene Atlas is an updateable transcriptome resource generated from a multi-laboratory collaboration following documented growth, sequencing and data analysis protocols for a number of evolutionarily distant JGI flagship plant species. With common comparable tissues and conditions across these plants, Gene Atlas provides insights into the changes in transcriptional landscape in response to conditions across diverse plant genomes. In addition to cataloging gene expression profiles this growing resource significantly contributes to the JGI genome annotation efforts to accurately annotate genetic elements.
Currently, JGI Gene Atlas consists of >2,000 RNA-seq samples from seventeen JGI plant flagship genomes: Chlamydomonas reinhardtii (algal model), Physcomitrella patens (moss model), Arabidopsis thaliana (model for plant genetics and biology), Sphagnum angustifolium (aka. S. fallax) (flat-topped bogmoss), Glycine max (legume model and crop plant), Medicago truncatula (forage legume model), Populus trichocarpa (woody perennial biomass crop), Eucalyptus grandis (biomass energy crop), Brachypodium distachyon (a C3 grass model), Panicum virgatum (herbaceous perennial crop), Panicum hallii var. filipes (grass model), Panicum hallii var. hallii (grass model), Setaria italica (grain and forage crop), Setaria viridis (a C4 grass model), Sorghum bicolor (a C4 grass bioenergy crop), Sorghum bicolor Rio (sweet sorghum, Rio) and Kalanchoe fedtschenkoi (model for Crassulacean acid metabolism photosynthesis). For each of these plants, samples were collected from coordinated studies including developmental stages in Physcomitrella and sorghum, abiotic stress conditions in Brachypodium and Kalanchoe and a common comparative condition across many representative plants focused on nitrogen metabolism. These dense RNA-seq data sets further allow to compare gene expression across conditions within a plant and through orthologous gene sets across phylogenetically diverse plant genomes.
This website provides a hub for access to the JGI Plant Gene Atlas data, subject to the data usage policy.
Citing JGI Plant Gene Atlas data
Sreedasyam,A., Plott,C., Hossain,M.S., Lovell,J.T., Grimwood,J., Jenkins,J.W., Daum,C., Barry,K., Carlson,J., Shu,S., et al. (2023) JGI Plant Gene Atlas: an updateable transcriptome resource to improve functional gene descriptions across the plant kingdom. Nucleic Acids Res., 10.1093/nar/gkad616..
Contacts
- Jeremy Schmutz (jschmutz AT hudsonalpha DOT org)
- Avinash Sreedasyam (asreedasyam AT hudsonalpha DOT org)
JGI, Integrative Genomics Building | 1 Cyclotron Dr., Building 91, Berkeley, CA 94720
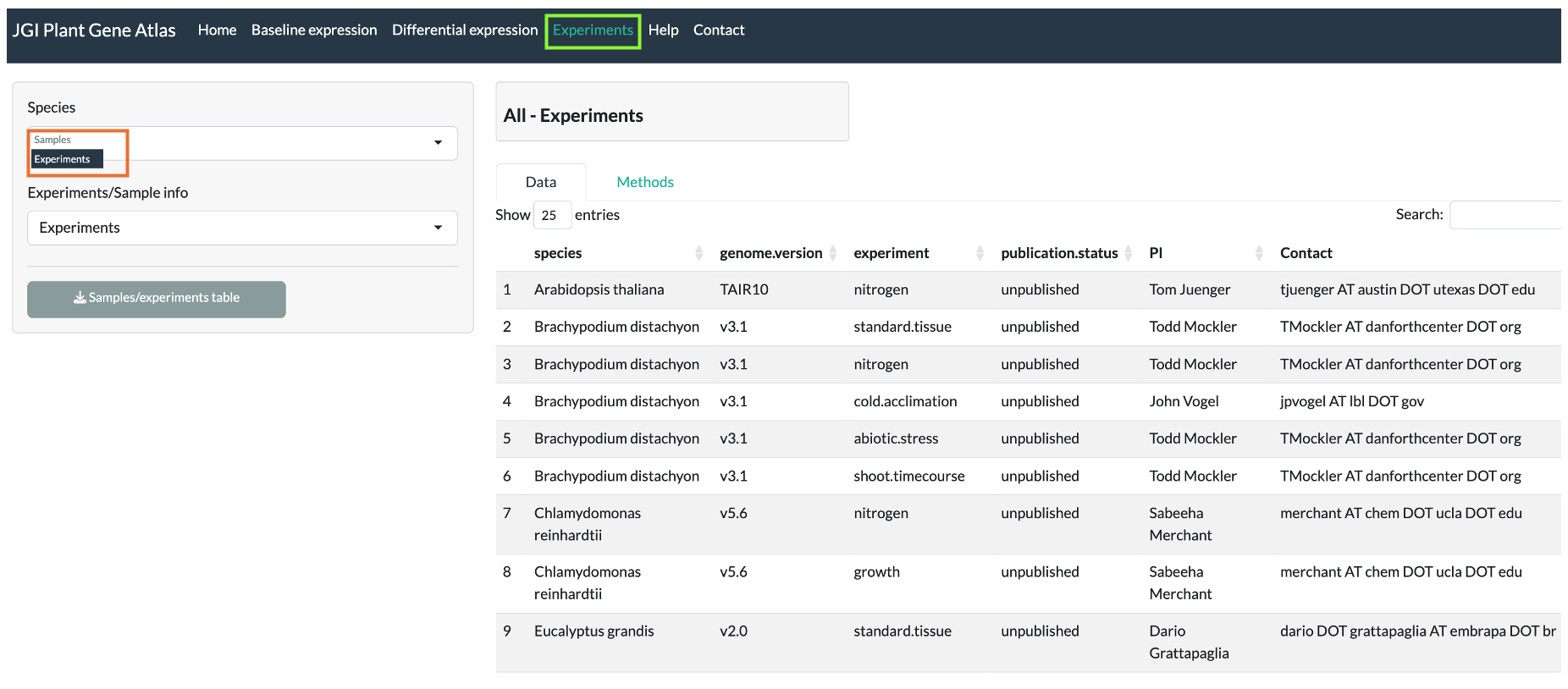
Gene Atlas experiments
Most common experiments includes but not limited to:
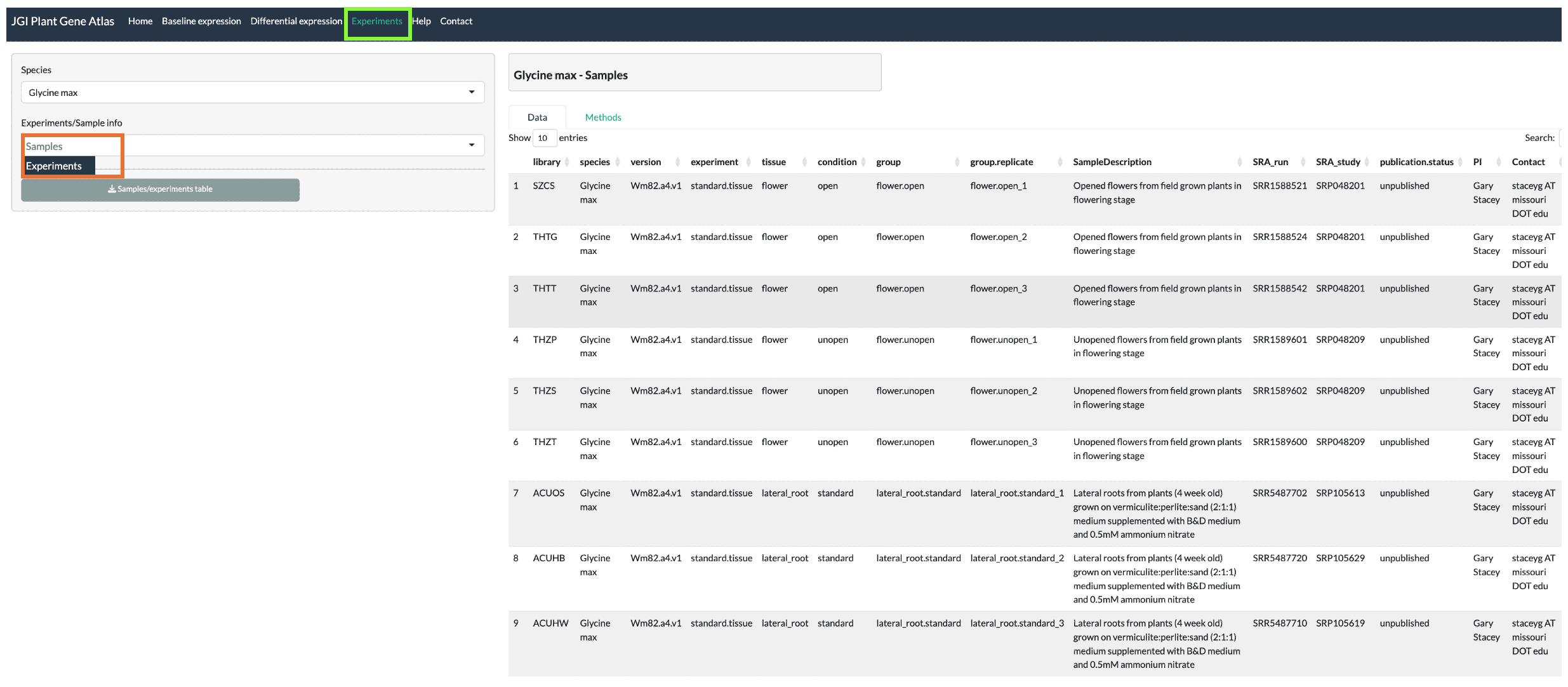
Standard tissues
Plants are grown in standard conditions in green house for four weeks and
leaf, stem, roots, lateral roots, buds, and flowers are harvested.
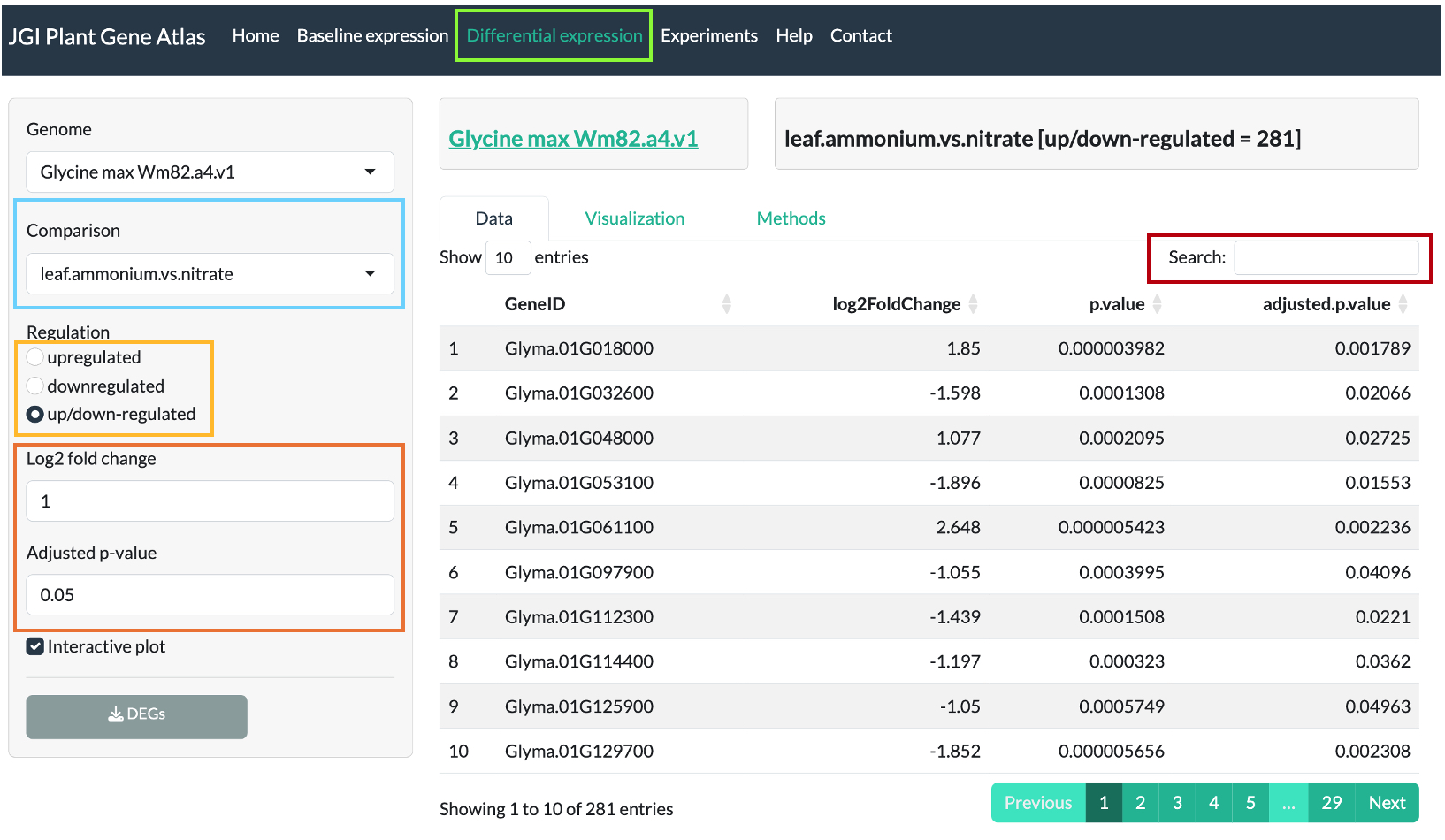
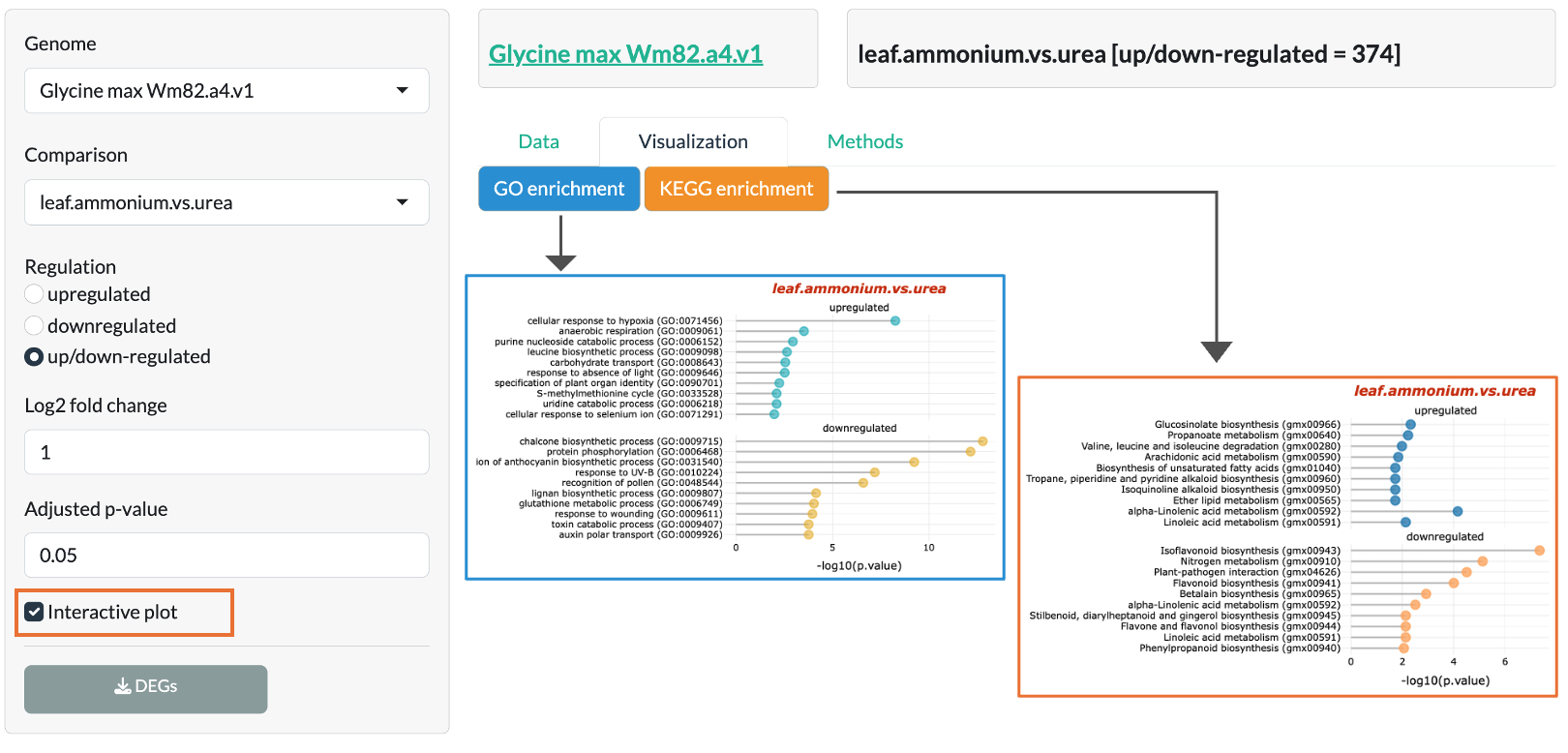
Nitrogen experiment
Plants for nitrogen experiment are grown in green house under standard conditions and
supplemented with nutrient solution containing either 10 mM KNO3 (nitrate treatment)
or 10 mM (NH4)3PO4 (ammonium treatment) or 10 mM urea (urea treatment). The nutrient solutions are renewed every 3 days and leaf, stem, and roots are harvested after 4 weeks.
Harvested tissues are immediately flash frozen in liquid nitrogen and stored at -80oC for RNA extraction.